Our Research
State of the Art
In contrast to Arabica, research and development activities on Robusta coffee are limited. Moreover, studies are usually performed in a small number of countries, such as Brazil, Vietnam, Indonesia, Uganda, and India. Research and development on Robusta is impeded by the restricted number of studies on the (wild) genetic diversity and limited access to genetic resources. By performing research in the DRC, a country of origin of coffee with important genetic diversity of wild Coffea canephora and a big potential for coffee cultivation, we contribute to local and international needs.
An in depth socio-economic study is being performed to ensure that all socio-economic, cultural-historic, and governmental considerations are understood. This will ascertain whether or not local populations can be encouraged to adopt coffee cultivation in their livelihood strategies. This will be done by carrying out an in-depth qualitative sociological analysis, assessing how potential beneficiaries perceive coffee cultivation. Next, we will determine how investment in coffee would impact other livelihood activities and what implications this would have for the environment. Then, the institutional interactions and social advantages and disadvantages will be analyzed to assess the governance framework in which the potential coffee production would take place. Lastly, behavioral changes will be documented over time.
Tasks:
- Inventory of the current coffee cultivation practices
- Assessment of the land ownership, territorial control, and land appropriation in the Tshopo region
- Current economic activities and interaction with markets
- Relations between farmers and state institutions
- Age and gender in coffee growing and related activities
- Investment and income in coffee growing
The diversity of the coffee genetic resources kept in the INERA Yangambi collection will be described and characterized in various different ways.
We aim to genetically screen the current Robusta collection at Yangambi using modern molecular techniques and compare it to wild collected material and commercially grown Robusta. The genetic passport data and subsequent population genetic network generated will act as a framework for data obtained in the other WP’s and will allow to carry out trait analyses.
For the SSR analyses, we use the microsatellite primers specifically designed by Poncet et al. (2007) and Hendre et al. (2008) to analyze Robusta populations, that are amplified and subsequently genotyped. The fragment size-based SSR approach is complementary to the GBS technique that generates several Mbp of SNPs. A set of GBS-based SNPs is subsequently selected to generate a multiplex amplicon sequencing to obtain at least 500 SNPs included in short fragments of maximum 200bp to also cover highly degraded material such as herbarium samples (used as a historical reference). Using the assay approach allows us to genotype cheaply and time-efficiently, delivering high-throughput screening. General population genetic indices are calculated such as number of alleles per locus (Na), gene diversity, observed (Ho) and expected heterozygosity (He), gene diversity, and allelic richness (AR) using Genetix 4.0 and SPaGeDi 1.5. Population structure within the germplasm collection is investigated with a multivariate method that identifies and designates clusters of related individuals: DAPC (discriminant analysis of principal components). A Bayesian clustering algorithm as implemented in Structure 2.3, is used to estimate the number of genetic clusters. Both SSRs and GBS-derived SNP data are examined using statistical methods. Obtained results from SNP and SSR data analysis are compared to obtain a reliable estimate of the genetic variation within the collection and compared to the wild accessions.
Tasks:
- Genetic screening of the Robusta collection, combining single sequence repeats (SSR) and genotyping by sequencing (GBS)
- Genetic analysis
The collection will also be analyzed phenotypically. Existing and newly introduced genetic lines will be characterized based on descriptors developed by the International Plant Genetic Resources Institute (IPGRI, 1996). Values for 34 qualitative and 20 quantitative descriptors are recorded for up to three plants per accession and analysed using UPGMA (unweighted pair group method with arithmetic mean) clustering and ordination techniques to differentiate phenotypic entities. These descriptors should enable easy and quick discrimination between phenotypes, but they also provide critical information for proper management of accessions.
- Optimization of the descriptor list for Robusta coffee and development of a database with descriptor information
- Completion of the descriptor list for all accessions currently in the collection and for newly collected wild coffee accessions
- Analysis and summarization of the descriptor list
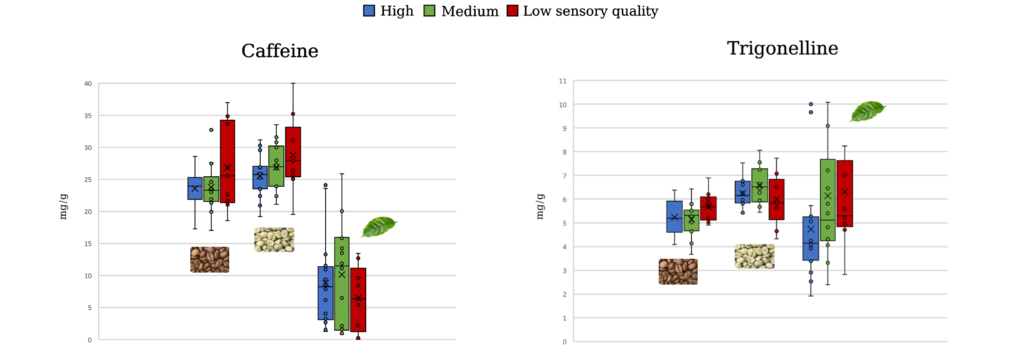
The chemical composition of coffee beans has been correlated with sensory quality. However, the leaves are often overlooked despite having unique metabolite profiles. An untargeted metabolomics study was performed relating metabolites in coffee leaves, green coffee beans, and roasted coffee beans to sensory quality and genetic background. Liquid chromatography – high-resolution mass spectrometry (LC-HRMS) was used to establish metabolite profiles for 39 Robusta coffee genotypes from the INERA Yangambi coffee collection. By using an untargeted metabolomics methodology developed by Montis et al. (2022), unbiased metabolite profiles were obtained that did not focus on specific compounds but rather looked at the total chemical composition of the plant material.
Interestingly, the metabolite profiles of coffee leaves were more accurate in predicting the sensory quality of a genotype than those of green or roasted beans. Several compounds were marked as potential predictors of low or high sensory quality in leaves, green beans, and roasted coffee beans. Further, the genetic background of the genotypes could only be predicted by leaf metabolite profiles, not by those in coffee beans. This predictive power of coffee leaf metabolite profiles could help accelerate the selection of genotypes with superior sensory quality. Interestingly, a wide range in green bean caffeine concentration was found within the studied genotypes (from 1.9 to 4.0%).
For more details, read the paper here.

Organoleptic (sensory) attributes are key determinants of coffee’s price and position in the global market. Therefore, the sensory profiles of 70 genetic lines from the collection were evaluated to assess their quality, commercial potential, and differences.
The coffee beans were picked and processed in a standardised manner, such that the results could be compared with the genetic, phenotypic, and chemical characteristics of the genetic lines, as well as with the agronomic characteristics of the sourced plants. Defect beans were removed and good-quality beans were bagged and coded by local partners. Using the Fine Robusta Standards and Protocols alongside the Coffee Taster’s Flavor Wheel, the coffees were evaluated by a panel of certified Q-graders and compared to commercial reference samples of graded quality Robusta.

The study uncovered several promising and unique sensory profiles in the collection, including descriptors such as Nutty/Cocoa, Fruity, Sweet, and Sour/Fermented. Some genotypes obtained scores up to 84.75, well above the threshold to be considered ‘Fine’ Robusta (equivalent to ‘Specialty’ in Arabica). Moreover, genetic lines seem to have similar sensory profiles over two harvest years, indicating consistent differences between genetic lines. These results show that the Robusta genetic diversity in the DR Congo can play an important role in coffee improvement, highlighting the importance of the INERA Yangambi collection.
For more info, take a look at the paper here.
To promote sustainable Robusta coffee production, the current coffee systems must first be assessed and characterized regarding their agro-ecological characteristics. The relationship between yield, biodiversity, and carbon stocks was evaluated in four coffee systems (coffee monocultures, cultivated agroforestry, wild agroforestry and forest coffee) to identify synergies and trade-offs between these metrics.
- Coffee yield in cultivated agroforestry does not differ significantly from that of coffee monocultures, despite the latter storing almost twice as much carbon.
- Trade-offs exist between woody plant diversity/carbon stocks and coffee yield.
- Co-benefits exist between woody plant diversity and carbon stocks.
- Cultivated coffee agroforestry systems are recommended over monocultures and wild coffee agroforestry from a perspective of balancing production, biodiversity, and carbon sequestration. However, considering the much higher woody plant diversity and carbon stocks, conserving forest coffee is of paramount importance.
Regarding soil fertility, soils were often found to be highly acidic (pH < 4,5). This could be combatted by liming the soil, improving the availability of nutrients and mitigating leaf nutrient deficiencies and imbalances. Additionally, yield differences are explained by differences in management, indicating that improving agricultural practices could significantly boost coffee yields.
For more information on the crop systems in the Tshopo province, read the paper here.
The successful introduction of Robusta coffee in the early 20th century as an alternative crop for Arabica coffee, plagued by coffee leaf rust, can be seen as an important milestone in the adaptation of coffee cultivation to a quickly emerging and devastating pest. Although it is widely accepted that Lucien Linden had an important role in the introduction of Robusta coffee and that he sourced it from the Congo, this history is not elucidated and many questions remain unanswered, e.g. how this exploration and introduction was organized, what is the exact origin of the cultivated genetic resources, which networks connected researchers traders in Belgian Congo and other colonial territories in Africa and Asia, etc. A better insight in the introduction and diffusion history of Robusta coffee and relevant international scientific networks, can help to answer actual questions in the context of in-situ and ex-situ conservation and for breeding towards a more climate resilient coffee cultivation, as current coffee cultivation is based on a very restricted genetic base mainly from the Congo Basin.
Tasks:
- Historical research: prospecting archival sources
- Digitizing and making documents available online
- Study of the collected information: compiling the information on the introduction, diffusion and further scientific valorization of Robusta coffee in Belgian Congo and other colonies in Africa and Asia (1880-1960)











